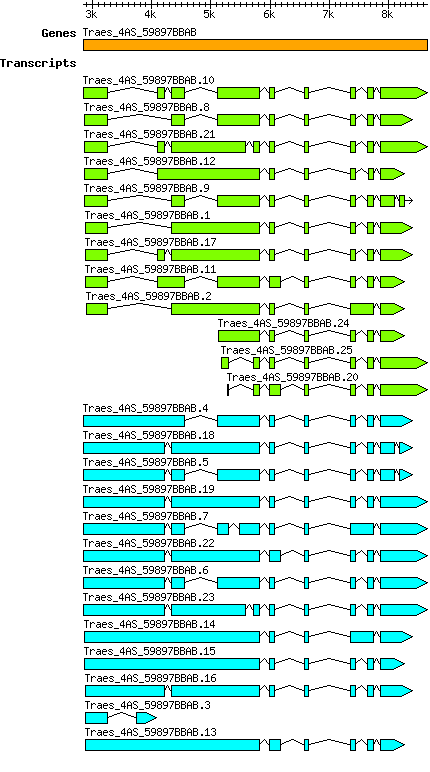
Gene
Taestivum Traes 4AS 59897BBAB
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_4AS_59897BBAB Traes_4AS_59897BBAB |
ta_iwgsc_4as_v2_5994705 | 2842 | 8665 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_4AS_59897BBAB.1 Traes_4AS_59897BBAB.1 PAC:32022796 |
High | No | 2732 | Get ORF Coding potential... Type: noncoding Potential: -1.179 Folding energies... MFEI: -29.046 AMFE: -0.646 GC content %: 44.985 | |
| Taestivum_Traes_4AS_59897BBAB.10 Traes_4AS_59897BBAB.10 PAC:32022805 |
High | No | 2586 | Get ORF Coding potential... Type: noncoding Potential: -1.178 Folding energies... MFEI: -32.660 AMFE: -0.661 GC content %: 49.381 | |
| Taestivum_Traes_4AS_59897BBAB.11 Traes_4AS_59897BBAB.11 PAC:32022806 |
High | No | 2377 | Get ORF Coding potential... Type: noncoding Potential: -1.175 Folding energies... MFEI: -30.665 AMFE: -0.661 GC content %: 46.403 | |
| Taestivum_Traes_4AS_59897BBAB.12 Traes_4AS_59897BBAB.12 PAC:32022807 |
High | No | 2839 | Get ORF Coding potential... Type: noncoding Potential: -1.180 Folding energies... MFEI: -28.733 AMFE: -0.649 GC content %: 44.241 | |
| Taestivum_Traes_4AS_59897BBAB.13 Traes_4AS_59897BBAB.13 PAC:32022808 |
Low | No | 3780 | Get ORF Coding potential... Type: noncoding Potential: -1.187 Folding energies... MFEI: -28.452 AMFE: -0.644 GC content %: 44.153 | |
| Taestivum_Traes_4AS_59897BBAB.14 Traes_4AS_59897BBAB.14 PAC:32022813 |
Low | No | 4050 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -28.332 AMFE: -0.643 GC content %: 44.074 | |
| Taestivum_Traes_4AS_59897BBAB.15 Traes_4AS_59897BBAB.15 PAC:32022811 |
Low | No | 3697 | Get ORF Coding potential... Type: noncoding Potential: -1.186 Folding energies... MFEI: -28.308 AMFE: -0.639 GC content %: 44.333 | |
| Taestivum_Traes_4AS_59897BBAB.16 Traes_4AS_59897BBAB.16 PAC:32022809 |
Low | No | 3713 | Get ORF Coding potential... Type: noncoding Potential: -1.186 Folding energies... MFEI: -28.805 AMFE: -0.643 GC content %: 44.789 | |
| Taestivum_Traes_4AS_59897BBAB.17 Traes_4AS_59897BBAB.17 PAC:32022814 |
High | No | 2864 | Get ORF Coding potential... Type: noncoding Potential: -1.180 Folding energies... MFEI: -29.268 AMFE: -0.653 GC content %: 44.797 | |
| Taestivum_Traes_4AS_59897BBAB.18 Traes_4AS_59897BBAB.18 PAC:32022810 |
Low | No | 3644 | Get ORF Coding potential... Type: noncoding Potential: -1.186 Folding energies... MFEI: -28.890 AMFE: -0.640 GC content %: 45.170 | |
| Taestivum_Traes_4AS_59897BBAB.19 Traes_4AS_59897BBAB.19 PAC:32022812 |
Low | No | 3998 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -30.004 AMFE: -0.649 GC content %: 46.223 | |
| Taestivum_Traes_4AS_59897BBAB.2 Traes_4AS_59897BBAB.2 PAC:32022797 |
High | No | 2786 | Get ORF Coding potential... Type: noncoding Potential: -1.180 Folding energies... MFEI: -28.304 AMFE: -0.646 GC content %: 43.826 | |
| Taestivum_Traes_4AS_59897BBAB.20 Traes_4AS_59897BBAB.20 PAC:32022815 |
High | No | 1354 | Get ORF Coding potential... Type: noncoding Potential: -1.071 Folding energies... MFEI: -32.925 AMFE: -0.662 GC content %: 49.705 | |
| Taestivum_Traes_4AS_59897BBAB.21 Traes_4AS_59897BBAB.21 PAC:32022818 |
High | No | 2986 | Get ORF Coding potential... Type: noncoding Potential: -1.061 Folding energies... MFEI: -30.797 AMFE: -0.654 GC content %: 47.086 | |
| Taestivum_Traes_4AS_59897BBAB.22 Traes_4AS_59897BBAB.22 PAC:32022816 |
Low | No | 4093 | Get ORF Coding potential... Type: noncoding Potential: -1.188 Folding energies... MFEI: -30.137 AMFE: -0.655 GC content %: 46.005 | |
| Taestivum_Traes_4AS_59897BBAB.23 Traes_4AS_59897BBAB.23 PAC:32022817 |
Low | No | 3850 | Get ORF Coding potential... Type: noncoding Potential: -1.078 Folding energies... MFEI: -30.190 AMFE: -0.646 GC content %: 46.701 | |
| Taestivum_Traes_4AS_59897BBAB.24 Traes_4AS_59897BBAB.24 PAC:32022819 |
High | No | 1432 | Get ORF Coding potential... Type: noncoding Potential: -1.172 Folding energies... MFEI: -28.010 AMFE: -0.659 GC content %: 42.528 | |
| Taestivum_Traes_4AS_59897BBAB.25 Traes_4AS_59897BBAB.25 PAC:32022820 |
High | No | 1346 | Get ORF Coding potential... Type: noncoding Potential: -0.927 Folding energies... MFEI: -31.642 AMFE: -0.639 GC content %: 49.480 | |
| Taestivum_Traes_4AS_59897BBAB.3 Traes_4AS_59897BBAB.3 PAC:32022798 |
Low | No | 705 | Get ORF Coding potential... Type: noncoding Potential: -1.105 Folding energies... MFEI: -33.574 AMFE: -0.596 GC content %: 56.312 | |
| Taestivum_Traes_4AS_59897BBAB.4 Traes_4AS_59897BBAB.4 PAC:32022799 |
Low | No | 3304 | Get ORF Coding potential... Type: noncoding Potential: -1.184 Folding energies... MFEI: -29.752 AMFE: -0.638 GC content %: 46.640 | |
| Taestivum_Traes_4AS_59897BBAB.5 Traes_4AS_59897BBAB.5 PAC:32022800 |
Low | No | 3090 | Get ORF Coding potential... Type: noncoding Potential: -1.182 Folding energies... MFEI: -30.324 AMFE: -0.642 GC content %: 47.249 | |
| Taestivum_Traes_4AS_59897BBAB.6 Traes_4AS_59897BBAB.6 PAC:32022802 |
Low | No | 3435 | Get ORF Coding potential... Type: noncoding Potential: -1.185 Folding energies... MFEI: -31.392 AMFE: -0.651 GC content %: 48.239 | |
| Taestivum_Traes_4AS_59897BBAB.7 Traes_4AS_59897BBAB.7 PAC:32022801 |
Low | No | 3453 | Get ORF Coding potential... Type: noncoding Potential: -1.113 Folding energies... MFEI: -31.014 AMFE: -0.647 GC content %: 47.958 | |
| Taestivum_Traes_4AS_59897BBAB.8 Traes_4AS_59897BBAB.8 PAC:32022803 |
High | No | 2191 | Get ORF Coding potential... Type: noncoding Potential: -1.173 Folding energies... MFEI: -30.895 AMFE: -0.644 GC content %: 47.969 | |
| Taestivum_Traes_4AS_59897BBAB.9 Traes_4AS_59897BBAB.9 PAC:32022804 |
High | No | 1946 | Get ORF Coding potential... Type: noncoding Potential: -1.168 Folding energies... MFEI: -31.141 AMFE: -0.646 GC content %: 48.201 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_4AS_59897BBAB.22 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.14 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.7 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.23 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.15 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.3 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.16 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.4 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.18 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV | |
| Taestivum_Traes_4AS_59897BBAB.5 | RepBase (RepeatMasker) | 2014-01-31 | EnSpm1_HV |
... further results
Gene models