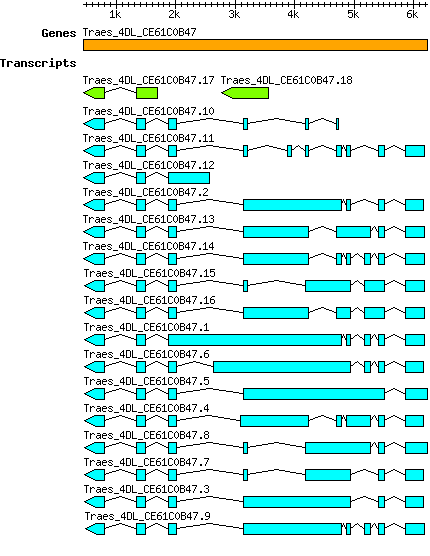
Gene
Taestivum Traes 4DL CE61C0B47
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_4DL_CE61C0B47 Traes_4DL_CE61C0B47 |
ta_iwgsc_4dl_v3_14362524 | 446 | 6232 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_4DL_CE61C0B47.1 Traes_4DL_CE61C0B47.1 PAC:31968302 |
Low | No | 3997 | Get ORF Coding potential... Type: noncoding Potential: -0.280 Folding energies... MFEI: -28.742 AMFE: -0.675 GC content %: 42.557 | |
| Taestivum_Traes_4DL_CE61C0B47.10 Traes_4DL_CE61C0B47.10 PAC:31968292 |
Low | No | 785 | Get ORF Coding potential... Type: noncoding Potential: -0.316 Folding energies... MFEI: -28.764 AMFE: -0.631 GC content %: 45.605 | |
| Taestivum_Traes_4DL_CE61C0B47.11 Traes_4DL_CE61C0B47.11 PAC:31968298 |
Low | No | 1400 | Get ORF Coding potential... Type: noncoding Potential: -0.180 Folding energies... MFEI: -31.479 AMFE: -0.623 GC content %: 50.500 | |
| Taestivum_Traes_4DL_CE61C0B47.12 Traes_4DL_CE61C0B47.12 PAC:31968293 |
Low | No | 1187 | Get ORF Coding potential... Type: noncoding Potential: -0.374 Folding energies... MFEI: -28.677 AMFE: -0.658 GC content %: 43.555 | |
| Taestivum_Traes_4DL_CE61C0B47.13 Traes_4DL_CE61C0B47.13 PAC:31968299 |
Low | No | 2709 | Get ORF Coding potential... Type: noncoding Potential: -0.219 Folding energies... MFEI: -29.417 AMFE: -0.646 GC content %: 45.515 | |
| Taestivum_Traes_4DL_CE61C0B47.14 Traes_4DL_CE61C0B47.14 PAC:31968300 |
Low | No | 2404 | Get ORF Coding potential... Type: noncoding Potential: -0.206 Folding energies... MFEI: -30.379 AMFE: -0.657 GC content %: 46.215 | |
| Taestivum_Traes_4DL_CE61C0B47.15 Traes_4DL_CE61C0B47.15 PAC:31968295 |
Low | No | 2120 | Get ORF Coding potential... Type: noncoding Potential: -0.232 Folding energies... MFEI: -29.491 AMFE: -0.627 GC content %: 47.028 | |
| Taestivum_Traes_4DL_CE61C0B47.16 Traes_4DL_CE61C0B47.16 PAC:31968297 |
Low | No | 2614 | Get ORF Coding potential... Type: noncoding Potential: -0.217 Folding energies... MFEI: -29.839 AMFE: -0.644 GC content %: 46.366 | |
| Taestivum_Traes_4DL_CE61C0B47.17 Traes_4DL_CE61C0B47.17 PAC:31968304 |
High | No | 698 | Get ORF Coding potential... Type: noncoding Potential: -1.149 Folding energies... MFEI: -26.562 AMFE: -0.618 GC content %: 42.980 | |
| Taestivum_Traes_4DL_CE61C0B47.18 Traes_4DL_CE61C0B47.18 PAC:31968303 |
High | No | 799 | Get ORF Coding potential... Type: noncoding Potential: -1.253 Folding energies... MFEI: -25.056 AMFE: -0.642 GC content %: 39.049 | |
| Taestivum_Traes_4DL_CE61C0B47.2 Traes_4DL_CE61C0B47.2 PAC:31968287 |
Low | No | 2760 | Get ORF Coding potential... Type: noncoding Potential: -0.232 Folding energies... MFEI: -29.370 AMFE: -0.670 GC content %: 43.841 | |
| Taestivum_Traes_4DL_CE61C0B47.3 Traes_4DL_CE61C0B47.3 PAC:31968288 |
Low | No | 2821 | Get ORF Coding potential... Type: noncoding Potential: -0.233 Folding energies... MFEI: -29.284 AMFE: -0.669 GC content %: 43.779 | |
| Taestivum_Traes_4DL_CE61C0B47.4 Traes_4DL_CE61C0B47.4 PAC:31968290 |
Low | No | 2652 | Get ORF Coding potential... Type: noncoding Potential: -0.221 Folding energies... MFEI: -29.713 AMFE: -0.651 GC content %: 45.664 | |
| Taestivum_Traes_4DL_CE61C0B47.5 Traes_4DL_CE61C0B47.5 PAC:31968289 |
Low | No | 3357 | Get ORF Coding potential... Type: noncoding Potential: -0.253 Folding energies... MFEI: -28.752 AMFE: -0.646 GC content %: 44.474 | |
| Taestivum_Traes_4DL_CE61C0B47.6 Traes_4DL_CE61C0B47.6 PAC:31968291 |
Low | No | 3429 | Get ORF Coding potential... Type: noncoding Potential: -0.261 Folding energies... MFEI: -29.058 AMFE: -0.675 GC content %: 43.074 | |
| Taestivum_Traes_4DL_CE61C0B47.7 Traes_4DL_CE61C0B47.7 PAC:31968301 |
Low | No | 1859 | Get ORF Coding potential... Type: noncoding Potential: -0.219 Folding energies... MFEI: -29.720 AMFE: -0.639 GC content %: 46.530 | |
| Taestivum_Traes_4DL_CE61C0B47.8 Traes_4DL_CE61C0B47.8 PAC:31968294 |
Low | No | 2252 | Get ORF Coding potential... Type: noncoding Potential: -0.237 Folding energies... MFEI: -29.284 AMFE: -0.628 GC content %: 46.625 | |
| Taestivum_Traes_4DL_CE61C0B47.9 Traes_4DL_CE61C0B47.9 PAC:31968296 |
Low | No | 2841 | Get ORF Coding potential... Type: noncoding Potential: -0.233 Folding energies... MFEI: -29.539 AMFE: -0.668 GC content %: 44.245 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_4DL_CE61C0B47.14 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.5 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.15 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.6 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.16 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.1 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.7 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.2 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.11 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM | |
| Taestivum_Traes_4DL_CE61C0B47.8 | RepBase (RepeatMasker) | 2014-01-31 | LATIDU1_TM |
... further results
Gene models