Gene
Taestivum Traes 7BL B56D80EF4
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_7BL_B56D80EF4 Traes_7BL_B56D80EF4 |
ta_iwgsc_7bl_v1_6658894 | 4 | 5744 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
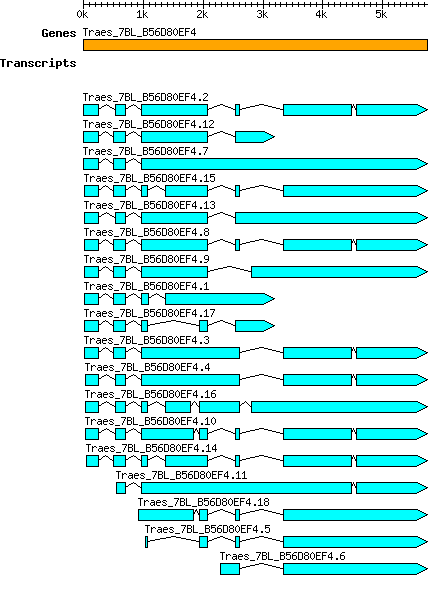
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_7BL_B56D80EF4.1 Traes_7BL_B56D80EF4.1 PAC:32020032 |
Low | No | 2355 | Get ORF Coding potential... Type: noncoding Potential: -0.119 Folding energies... MFEI: -28.166 AMFE: -0.687 GC content %: 41.019 | |
| Taestivum_Traes_7BL_B56D80EF4.10 Traes_7BL_B56D80EF4.10 PAC:32020026 |
Low | No | 3780 | Get ORF Coding potential... Type: noncoding Potential: -0.099 Folding energies... MFEI: -27.169 AMFE: -0.681 GC content %: 39.894 | |
| Taestivum_Traes_7BL_B56D80EF4.11 Traes_7BL_B56D80EF4.11 PAC:32020027 |
Low | No | 4843 | Get ORF Coding potential... Type: noncoding Potential: -0.114 Folding energies... MFEI: -26.042 AMFE: -0.693 GC content %: 37.601 | |
| Taestivum_Traes_7BL_B56D80EF4.12 Traes_7BL_B56D80EF4.12 PAC:32020025 |
Low | No | 2207 | Get ORF Coding potential... Type: noncoding Potential: -0.054 Folding energies... MFEI: -28.532 AMFE: -0.690 GC content %: 41.368 | |
| Taestivum_Traes_7BL_B56D80EF4.13 Traes_7BL_B56D80EF4.13 PAC:32020028 |
Low | No | 4724 | Get ORF Coding potential... Type: noncoding Potential: -0.113 Folding energies... MFEI: -26.763 AMFE: -0.686 GC content %: 39.035 | |
| Taestivum_Traes_7BL_B56D80EF4.14 Traes_7BL_B56D80EF4.14 PAC:32020030 |
Low | No | 3610 | Get ORF Coding potential... Type: noncoding Potential: -0.044 Folding energies... MFEI: -26.895 AMFE: -0.674 GC content %: 39.917 | |
| Taestivum_Traes_7BL_B56D80EF4.15 Traes_7BL_B56D80EF4.15 PAC:32020029 |
Low | No | 3725 | Get ORF Coding potential... Type: noncoding Potential: -0.048 Folding energies... MFEI: -27.052 AMFE: -0.674 GC content %: 40.161 | |
| Taestivum_Traes_7BL_B56D80EF4.16 Traes_7BL_B56D80EF4.16 PAC:32020031 |
Low | No | 4514 | Get ORF Coding potential... Type: noncoding Potential: -0.060 Folding energies... MFEI: -27.131 AMFE: -0.687 GC content %: 39.477 | |
| Taestivum_Traes_7BL_B56D80EF4.17 Traes_7BL_B56D80EF4.17 PAC:32020033 |
Low | No | 1321 | Get ORF Coding potential... Type: noncoding Potential: -0.197 Folding energies... MFEI: -29.932 AMFE: -0.684 GC content %: 43.755 | |
| Taestivum_Traes_7BL_B56D80EF4.18 Traes_7BL_B56D80EF4.18 PAC:32020034 |
Low | No | 3511 | Get ORF Coding potential... Type: noncoding Potential: -1.245 Folding energies... MFEI: -26.505 AMFE: -0.685 GC content %: 38.707 | |
| Taestivum_Traes_7BL_B56D80EF4.2 Traes_7BL_B56D80EF4.2 PAC:32020023 |
Low | No | 3918 | Get ORF Coding potential... Type: noncoding Potential: -0.100 Folding energies... MFEI: -26.986 AMFE: -0.680 GC content %: 39.689 | |
| Taestivum_Traes_7BL_B56D80EF4.3 Traes_7BL_B56D80EF4.3 PAC:32020021 |
Low | No | 4389 | Get ORF Coding potential... Type: noncoding Potential: -0.109 Folding energies... MFEI: -26.995 AMFE: -0.685 GC content %: 39.417 | |
| Taestivum_Traes_7BL_B56D80EF4.4 Traes_7BL_B56D80EF4.4 PAC:32020020 |
Low | No | 4351 | Get ORF Coding potential... Type: noncoding Potential: -0.109 Folding energies... MFEI: -26.766 AMFE: -0.683 GC content %: 39.209 | |
| Taestivum_Traes_7BL_B56D80EF4.5 Traes_7BL_B56D80EF4.5 PAC:32020017 |
Low | No | 2635 | Get ORF Coding potential... Type: noncoding Potential: -1.219 Folding energies... MFEI: -25.852 AMFE: -0.672 GC content %: 38.482 | |
| Taestivum_Traes_7BL_B56D80EF4.6 Traes_7BL_B56D80EF4.6 PAC:32020018 |
Low | No | 2710 | Get ORF Coding potential... Type: noncoding Potential: -1.222 Folding energies... MFEI: -25.122 AMFE: -0.668 GC content %: 37.601 | |
| Taestivum_Traes_7BL_B56D80EF4.7 Traes_7BL_B56D80EF4.7 PAC:32020024 |
Low | No | 5222 | Get ORF Coding potential... Type: noncoding Potential: -0.120 Folding energies... MFEI: -26.875 AMFE: -0.689 GC content %: 38.989 | |
| Taestivum_Traes_7BL_B56D80EF4.8 Traes_7BL_B56D80EF4.8 PAC:32020019 |
Low | No | 3940 | Get ORF Coding potential... Type: noncoding Potential: -0.100 Folding energies... MFEI: -27.079 AMFE: -0.681 GC content %: 39.772 | |
| Taestivum_Traes_7BL_B56D80EF4.9 Traes_7BL_B56D80EF4.9 PAC:32020022 |
Low | No | 4475 | Get ORF Coding potential... Type: noncoding Potential: -0.110 Folding energies... MFEI: -27.050 AMFE: -0.685 GC content %: 39.486 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_7BL_B56D80EF4.2 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.13 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.8 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.3 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.14 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.9 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.4 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.15 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.5 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe | |
| Taestivum_Traes_7BL_B56D80EF4.16 | RepBase (RepeatMasker) | 2014-01-31 | SINE2-1_TAe |
... further results
Gene models