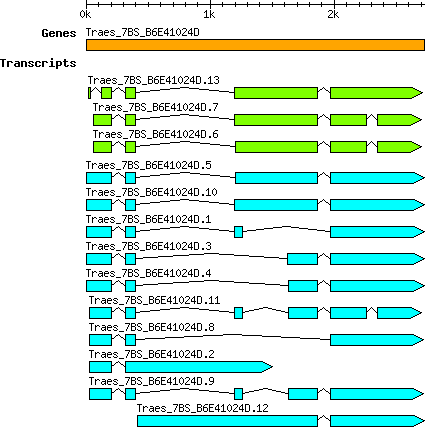
Gene
Taestivum Traes 7BS B6E41024D
From GreeNC
Gene information
| Gene alias / name | Chrom. | Start | End | Source | Assembly | Species | Details
|
|---|---|---|---|---|---|---|---|
| Taestivum_Traes_7BS_B6E41024D Traes_7BS_B6E41024D |
ta_iwgsc_7bs_v1_3154146 | 4 | 2725 | Phytozome | v2.2 | Triticum_aestivum | Coding? No |
Transcript features
This table shows how a lncRNA has been classified (high/low confidence) or whether it has been classified as pre-miRNA (go to FAQ for classification criteria). Besides, it displays the lncRNA length and sequence as well as information about the ORF, the coding potential calculated by the Coding Potential Calculator (CPC), and the folding energies calculated by RNAfold.
| Transcript alias / name | Confidence | Pre-miRNA | Length | Sequence | Details
|
|---|---|---|---|---|---|
| Taestivum_Traes_7BS_B6E41024D.1 Traes_7BS_B6E41024D.1 PAC:31972868 |
Low | No | 1104 | Get ORF Coding potential... Type: noncoding Potential: -1.313 Folding energies... MFEI: -28.234 AMFE: -0.604 GC content %: 46.739 | |
| Taestivum_Traes_7BS_B6E41024D.10 Traes_7BS_B6E41024D.10 PAC:31972867 |
Low | No | 1706 | Get ORF Coding potential... Type: noncoding Potential: -1.338 Folding energies... MFEI: -27.960 AMFE: -0.634 GC content %: 44.080 | |
| Taestivum_Traes_7BS_B6E41024D.11 Traes_7BS_B6E41024D.11 PAC:31972865 |
Low | No | 1212 | Get ORF Coding potential... Type: noncoding Potential: -1.319 Folding energies... MFEI: -29.373 AMFE: -0.620 GC content %: 47.360 | |
| Taestivum_Traes_7BS_B6E41024D.12 Traes_7BS_B6E41024D.12 PAC:31972869 |
Low | Yes | 2204 | Get ORF Coding potential... Type: noncoding Potential: -1.349 Folding energies... MFEI: -29.047 AMFE: -0.701 GC content %: 41.425 | |
| Taestivum_Traes_7BS_B6E41024D.13 Traes_7BS_B6E41024D.13 PAC:31972870 |
High | No | 1595 | Get ORF Coding potential... Type: noncoding Potential: -1.335 Folding energies... MFEI: -28.088 AMFE: -0.629 GC content %: 44.639 | |
| Taestivum_Traes_7BS_B6E41024D.2 Traes_7BS_B6E41024D.2 PAC:31972858 |
Low | Yes | 1367 | Get ORF Coding potential... Type: noncoding Potential: -1.400 Folding energies... MFEI: -26.474 AMFE: -0.704 GC content %: 37.601 | |
| Taestivum_Traes_7BS_B6E41024D.3 Traes_7BS_B6E41024D.3 PAC:31972859 |
Low | No | 1274 | Get ORF Coding potential... Type: noncoding Potential: -1.322 Folding energies... MFEI: -29.419 AMFE: -0.631 GC content %: 46.625 | |
| Taestivum_Traes_7BS_B6E41024D.4 Traes_7BS_B6E41024D.4 PAC:31972860 |
Low | No | 1271 | Get ORF Coding potential... Type: noncoding Potential: -1.322 Folding energies... MFEI: -29.237 AMFE: -0.628 GC content %: 46.577 | |
| Taestivum_Traes_7BS_B6E41024D.5 Traes_7BS_B6E41024D.5 PAC:31972862 |
Low | No | 1701 | Get ORF Coding potential... Type: noncoding Potential: -1.315 Folding energies... MFEI: -28.025 AMFE: -0.636 GC content %: 44.092 | |
| Taestivum_Traes_7BS_B6E41024D.6 Traes_7BS_B6E41024D.6 PAC:31972861 |
High | No | 1542 | Get ORF Coding potential... Type: noncoding Potential: -1.304 Folding energies... MFEI: -28.554 AMFE: -0.644 GC content %: 44.358 | |
| Taestivum_Traes_7BS_B6E41024D.7 Traes_7BS_B6E41024D.7 PAC:31972864 |
High | No | 1546 | Get ORF Coding potential... Type: noncoding Potential: -1.333 Folding energies... MFEI: -28.499 AMFE: -0.642 GC content %: 44.373 | |
| Taestivum_Traes_7BS_B6E41024D.8 Traes_7BS_B6E41024D.8 PAC:31972863 |
Low | No | 1015 | Get ORF Coding potential... Type: noncoding Potential: -1.307 Folding energies... MFEI: -28.296 AMFE: -0.605 GC content %: 46.798 | |
| Taestivum_Traes_7BS_B6E41024D.9 Traes_7BS_B6E41024D.9 PAC:31972866 |
Low | No | 1306 | Get ORF Coding potential... Type: noncoding Potential: -1.324 Folding energies... MFEI: -29.035 AMFE: -0.622 GC content %: 46.708 |
Matches to external databases
| Transcript alias | Database | Version | Hit | Evalue |
|---|---|---|---|---|
| Taestivum_Traes_7BS_B6E41024D.12 | RepBase (RepeatMasker) | 2014-01-31 | HADES_TA | |
| Taestivum_Traes_7BS_B6E41024D.12 | Rfam | 11.0 | MIR1122 | 2.2e-15 |
| Taestivum_Traes_7BS_B6E41024D.9 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad | |
| Taestivum_Traes_7BS_B6E41024D.2 | RepBase (RepeatMasker) | 2014-01-31 | HADES_TA | |
| Taestivum_Traes_7BS_B6E41024D.2 | Rfam | 11.0 | MIR1122 | 1.3e-15 |
| Taestivum_Traes_7BS_B6E41024D.3 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad | |
| Taestivum_Traes_7BS_B6E41024D.1 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad | |
| Taestivum_Traes_7BS_B6E41024D.4 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad | |
| Taestivum_Traes_7BS_B6E41024D.10 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad | |
| Taestivum_Traes_7BS_B6E41024D.5 | RepBase (RepeatMasker) | 2014-01-31 | MuDR-1_Mad |
... further results
Gene models